Mission: We propose to develop a comprehensive chromatin imaging atlas of peripheral blood mononuclear cells (PBMCs) in aging related human diseases. PBMCs circulate throughout the body and serve as ultra-sensitive detectors of diseases. When sensing disease signals, PBMCs activate various signaling pathways, ultimately mounting an immune response. We demonstrated in ongoing work and in initial successful clinical trials that these changes in PBMC state are associated with subtle alterations in 3D chromatin organization to regulate gene expression (1-10). The goal of this project is to develop an atlas of PBMCs using light microscopy combined with AI methods to characterize the subtle alterations in chromatin structure associated with different aging related diseases. Such an imaging atlas will provide new avenues for early diagnostics and tracking of therapeutic interventions.
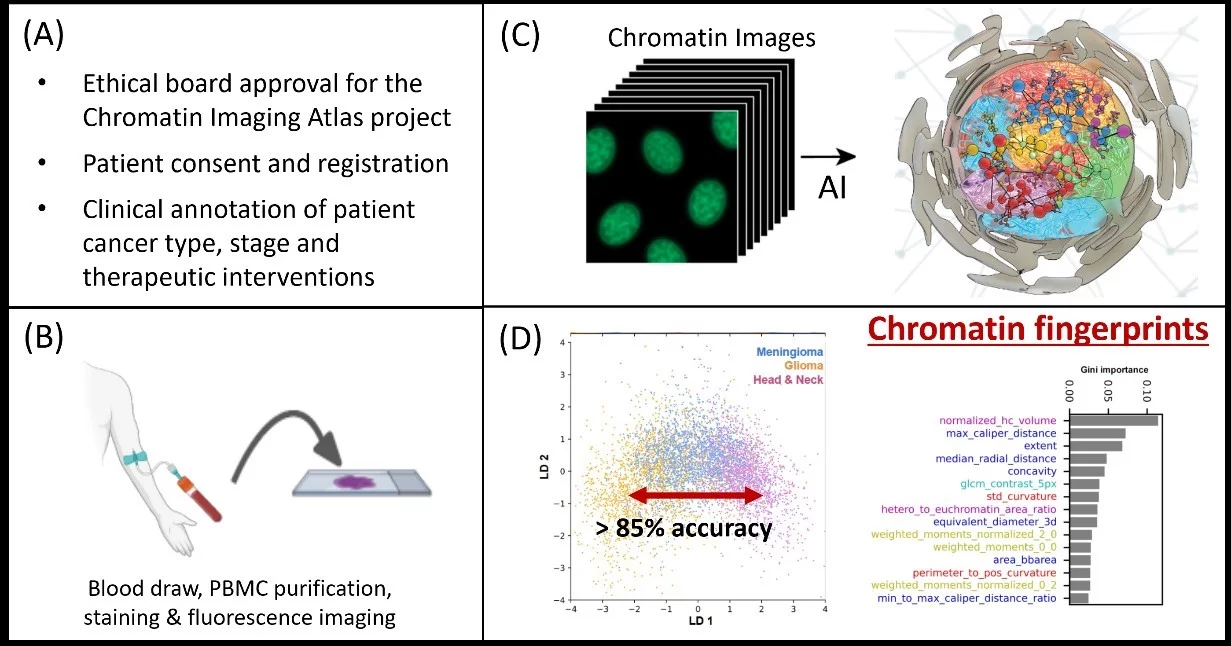
The four steps in establishing the Chromatin Imaging Atlas, which will initially comprise of N=10,000 patients with five major cancer types (Breast, Prostate, Lung, Colon, Head and Neck) and an equal number of healthy volunteers matching the patient demographics. (A) At the medical center, the ethical board approval, patient consent, and registration for the atlas project is obtained. In addition, the clinical parameters and history including cancer type, stage, and therapeutic interventions, with tissue biopsies, are annotated, and patient demographic data is obtained. (B) From a routine blood draw, peripheral blood mononuclear cells are purified, stained with chromatin markers, and imaged using an automated high-resolution fluorescence microscope. We will also implement the imaging on a microfluidic mobile microscope to enlarge the atlas through community screening and for providing point-of-care diagnostics. (C) Using our machine learning platform, the images are analyzed to obtain interpretable chromatin fingerprints. (D) This data will serve as a global reference map for research, for personalized and precision medicine, as well as in community screening programs for early diagnosis of different cancer types.
Implementation: To begin with, we will focus on human cancers and then extend this program to include metabolic and neurodegenerative disorders. With appropriate patient consent, the data will be generated as part of an international collaboration between medical centers across continents. Furthermore, we will expand the atlas by enabling community screening programs. Towards this, we will develop a microfluidic mobile phone microscope and apply it in large-scale field trials. The resulting images and AI-based chromatin fingerprints will be made available as open-access resources for researchers and clinicians. The proposed atlas will provide a reference map of PBMCs in the normal state and across aging related diseases for early diagnostics as well as for evaluating therapeutic options in personalized and precision medicine.
References:
1. A.Radhakrishnan, K.Damodharan, AC. Soylemezoglu, C.Uhler & G.V.Shivashankar, (2017). Machine learning for nuclear mechano-morphometric biomarkers in cancer diagnosis, Scientific Reports 7(1):17946
2. C.Uhler & G.V.Shivashankar, (2018). Nuclear mechanopathology & cancer diagnostics, Trends in Cancer, 4:320-331.
3. K.Damodharan, M.Cresenti, DS,Jokhun & G.V.Shivashankar, (2019). Nuclear morphometrics and chromatin condensation patterns as disease biomarkers using a mobile microscope. PloS One, 14(7), e0218757.
4. KD.Yang, K.Damodaran, S.Venkatachalapathy, AC.Soylemezoglu, G.V.Shivashankar & C.Uhler. (2020). Predicting cell lineages using autoencoders and optimal transport. PLOS Computational Biology, 16(4), 1-20.
5. KD.Yang, A.Belyaeva, S.Venkatachalapathy, K.Damodaran, A.Radhakrishnan, A.Katcoff, G.V.Shivashankar & C.Uhler, (2021). Multi-Domain Translation between Single-Cell Imaging and Sequencing Data using Autoencoders. Nature Communications, 12(1), 1-10.
6. S.Venkatachalapathy, DS.Jokhun, M.Andhari & G.V.Shivashankar, (2021). Single cell imaging-based chromatin biomarkers for tumor progression. Scientific Reports, 11(1), 1-14.
7. X.Zhang, X.Wang, G.V.Shivashankar & C.Uhler, (2022). Integration of Spatial Transcriptomics with Chromatin Images Using Graph-Based Autoencoder Identifies Joint Biomarkers for Alzheimer’s Disease. Nature Communications 13, 7480
8. C.Uhler & G.V.Shivashankar, (2022). Machine Learning approaches to Single-Cell Data Integration and Translation. Proceedings of the IEEE, 110(5), 557-576.
9. T.Pekec, S.Venkatachalpathy, M.Grzmil, R.Schibli, M.Behe & G.V.Shivashankar, (2023). Detecting radio- and chemoresistant cells in 3D cancer co-cultures using chromatin biomarkers. Scientific Reports 13(1), 20662.
10. K.Challa, D.Paysan, D.Leiser, N.Sauder, D.Weber & G.V.Shivashankar, (2023). Single-cell imaging-AI based chromatin biomarkers for diagnosis and therapy evaluation in tumor patients using liquid biopsies. npj Precision Oncology, 7(1), 135.
Some press releases about our recent paper: (https://www.nature.com/articles/s41698-023-00484-8)
https://www.psi.ch/en/media/our-research/enabling-early-detection-of-cancer
https://www.admin.ch/gov/en/start/documentation/media-releases.msg-id-99433.html
https://www.20min.ch/story/schweiz-durchbruch-bei-frueherkennung-von-krebs-952759521948
https://www.tagesanzeiger.ch/schweizer-forscher-durchbruch-bei-frueherkennung-von-krebs-585394277564

