Research Interests
All living cells are enveloped by a lipid bilayer called the cell membrane. The cell membrane controls the transport of chemicals and signals from the cytoplasm to the extracellular space, and vice versa, using specific membrane proteins embedded in the lipid bilayer. We are interesting on the structural aspects of channels and receptors and study them by diffraction methods using 3D and 2D crystals.
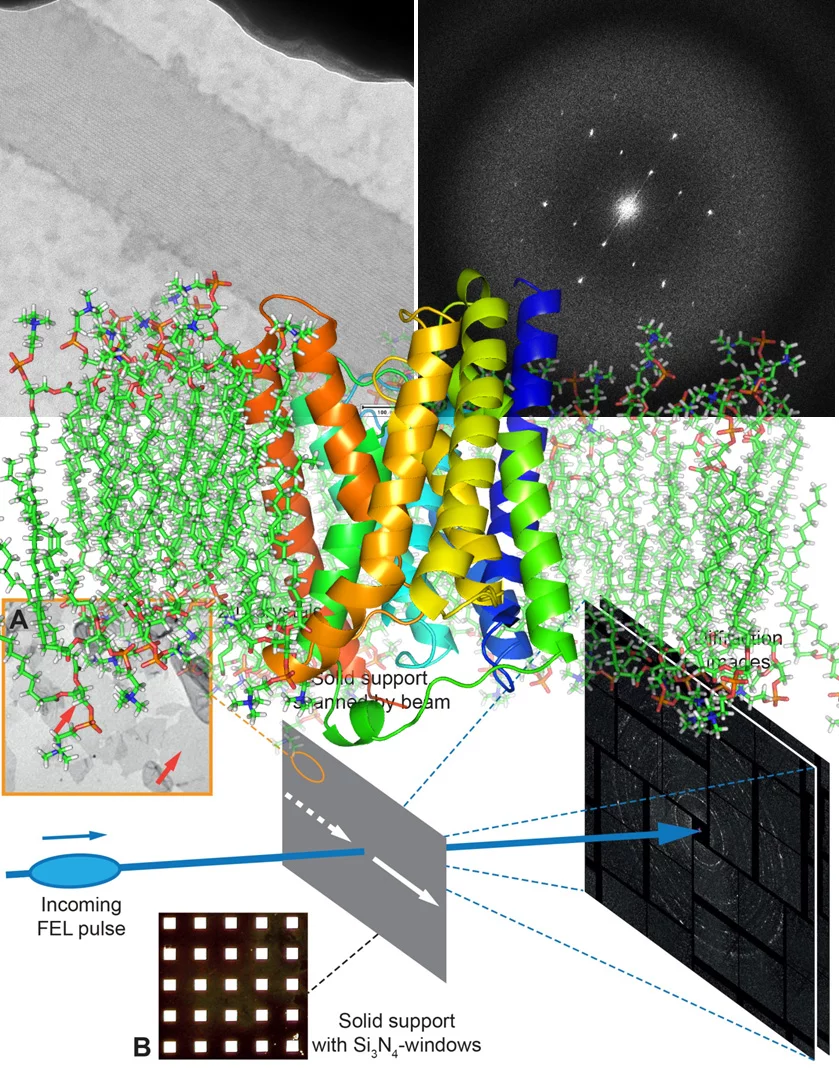
2D membrane protein crystals grow in the lipid bilayer and form mainly by hydrophobic interactions. 2D crystals have the advantage that the lipid-associated conformations of the membrane protein, due to the no crystal packing at the z-direction, therefore unlikely to adapt conformations which is less-like biological relevant. In addition, ligand with hydrophobic properties can free diffusion within the membrane bilayer. Freedom on the third dimension such as open and closed state of an ion channel can be detected, analyzed and compared to 3D structures obtained in the absence of lipids. Small size 2D-nanocrystals are easily obtained in the presence of ligands, pharmaceutical agents and regulatory nannobodies. The obtained structures of such membrane proteins will provide an important framework for functional characterization and for the identification of potential drug target sites including allosteric sides embedded into the lipid bilayer. We have made progress in terms of obtaining first 3D-structure of 2D crystals of a voltage-gated channel using cryo-EM and 4Å diffraction images from bacteriorhodopsin 2D crystals using X-ray free-lectron laser at LCLS, USA. 2D@FEL will allow studying the dynamic of the membrane protein in lipid bilayer at room temperature.
Our structural studies are complemented by biochemical and genetic analysis. We are collaborating with scientists from academia and from industry at the national and the international level. Being at the Paul Scherrer Institut, the future-host for SwissFEL, we are actively participating in the development of cutting-edge technologies for studying dynamics of membrane proteins using XFEL.
2D membrane protein crystals grow in the lipid bilayer and form mainly by hydrophobic interactions. 2D crystals have the advantage that the lipid-associated conformations of the membrane protein, due to the no crystal packing at the z-direction, therefore unlikely to adapt conformations which is less-like biological relevant. In addition, ligand with hydrophobic properties can free diffusion within the membrane bilayer. Freedom on the third dimension such as open and closed state of an ion channel can be detected, analyzed and compared to 3D structures obtained in the absence of lipids. Small size 2D-nanocrystals are easily obtained in the presence of ligands, pharmaceutical agents and regulatory nannobodies. The obtained structures of such membrane proteins will provide an important framework for functional characterization and for the identification of potential drug target sites including allosteric sides embedded into the lipid bilayer. We have made progress in terms of obtaining first 3D-structure of 2D crystals of a voltage-gated channel using cryo-EM and 4Å diffraction images from bacteriorhodopsin 2D crystals using X-ray free-lectron laser at LCLS, USA. 2D@FEL will allow studying the dynamic of the membrane protein in lipid bilayer at room temperature.
Our structural studies are complemented by biochemical and genetic analysis. We are collaborating with scientists from academia and from industry at the national and the international level. Being at the Paul Scherrer Institut, the future-host for SwissFEL, we are actively participating in the development of cutting-edge technologies for studying dynamics of membrane proteins using XFEL.
Group Members
Former Members
Research scientist
GPCR signaling :: membrane protein biochemistry
Publications
2021
-
Choo JPS, Kammerer RA, Li X, Li Z
High-Level Production of Phenylacetaldehyde using Fusion-Tagged Styrene Oxide Isomerase
Advanced Synthesis and Catalysis. 2021; 363(6): 1714-1721. https://doi.org/10.1002/adsc.202001500
DORA PSI -
Leka O, Wu Y, Li X, Kammerer RA
Crystal structure of the catalytic domain of botulinum neurotoxin subtype A3
Journal of Biological Chemistry. 2021; 296: 100684 (8 pp.). https://doi.org/10.1016/j.jbc.2021.100684
DORA PSI
2020
-
Li X, Brunner C, Wu Y, Leka O, Schneider G, Kammerer RA
Structural insights into the interaction of botulinum neurotoxin a with its neuronal receptor SV2C
Toxicon. 2020; 175: 36-43. https://doi.org/10.1016/j.toxicon.2019.11.010
DORA PSI -
Mortelmans T, Kazazis D, Guzenko VA, Padeste C, Braun T, Li X, et al.
Grayscale e-beam lithography: effects of a delayed development for well-controlled 3D patterning
Microelectronic Engineering. 2020; 225: 111272 (5 pp.). https://doi.org/10.1016/j.mee.2020.111272
DORA PSI
2019
-
Casadei CM, Nass K, Barty A, Hunter MS, Padeste C, Tsai C-J, et al.
Structure-factor amplitude reconstruction from serial femtosecond crystallography of two-dimensional membrane-protein crystals
IUCrJ. 2019; 6: 34-45. https://doi.org/10.1107/S2052252518014641
DORA PSI -
Górzny MŁ, Opara NL, Guzenko VA, Cadarso VJ, Schift H, Li XD, et al.
Microfabricated silicon chip as lipid membrane sample holder for serial protein crystallography
Micro and Nano Engineering. 2019; 3: 31-36. https://doi.org/10.1016/j.mne.2019.03.002
DORA PSI
2018
- Resolution extension by image summing in serial femtosecond crystallography of two-dimensional membrane-protein crystals
IUCRJ 5, 103 (2018).DOI: 10.1107/S2052252517017043
2017
-
Crystal structure of the BoNT/A2 receptor-binding domain in complex with the luminal domain of its neuronal receptor SV2C
Scientific Reports 7, 43588 (2017).DOI: 10.1038/srep43588
2015
- Large scale expression and purification of the rat 5-HT2c receptor
PROTEIN EXPRESSION AND PURIFICATION 106, 1 (2015).DOI: 10.1016/j.pep.2014.10.010
- Low-Z polymer sample supports for fixed-target serial femtosecond X-ray crystallography
JOURNAL OF APPLIED CRYSTALLOGRAPHY 48, 1072-1079 (2015).DOI: 10.1107/S1600576715010493
- Time-resolved structural studies with serial crystallography: A new light on retinal proteins
STRUCTURAL DYNAMICS 2, 041718 (2015).DOI: 10.1063/1.4922774
2014
- 7 angstrom resolution in protein two-dimensional-crystal X-ray diffraction at Linac Coherent Light Source
PHILOSOPHICAL TRANSACTIONS OF THE ROYAL SOCIETY B-BIOLOGICAL SCIENCES 369, UNSP 20130500 (2014).DOI: 10.1098/rstb.2013.0500
- A Type IV Translocated Legionella Cysteine Phytase Counteracts Intracellular Growth Restriction by Phytate
JOURNAL OF BIOLOGICAL CHEMISTRY 289, 34175-34188 (2014).DOI: 10.1074/jbc.M114.592568
- Femtosecond X-ray diffraction from two-dimensional protein crystals
IUCRJ 1, 95-100 (2014).DOI: 10.1107/S2052252514001444
2013
- Two Alternative Conformations of a Voltage-Gated Sodium Channel
JOURNAL OF MOLECULAR BIOLOGY 425, 4074 (2013).DOI: 10.1016/j.jmb.2013.06.036
2011
- Crystal structure of the GlnZ-DraG complex reveals a different form of P-II-target interaction
PROCEEDINGS OF THE NATIONAL ACADEMY OF SCIENCES OF THE UNITED STATES OF AMERICA 108, 18972 (2011).DOI: 10.1073/pnas.1108038108
- Quaternary Structure of the Oxaloacetate Decarboxylase Membrane Complex and Mechanistic Relationships to Pyruvate Carboxylases
JOURNAL OF BIOLOGICAL CHEMISTRY 286, 9457 (2011).DOI: 10.1074/jbc.M110.197442
2010
- A New P-II Protein Structure Identifies the 2-Oxoglutarate Binding Site
JOURNAL OF MOLECULAR BIOLOGY 400, 531 (2010).DOI: 10.1016/j.jmb.2010.05.036
2008
- Neuropilin-1 in regulation of VEGF-induced activation of p38MAPK and endothelial cell organization
BLOOD 112, 3638 (2008).DOI: 10.1182/blood-2007-12-125856
- Orf virus VEGF-E NZ2 promotes paracellular NRP-1/VEGFR-2 coreceptor assembly via the peptide RPPR
FASEB JOURNAL 22, 3078 (2008).DOI: 10.1096/fj.08-107219
2007
- Structural and mechanistic aspects of Amt/Rh proteins
JOURNAL OF STRUCTURAL BIOLOGY 158, 472 (2007).DOI: 10.1016/j.jsb.2007.01.004
- The 1.3-A resolution structure of nitrosomas erropea Rh50 and mechanistic implications for NH3 transport by Rhesus family proteins
PROCEEDINGS OF THE NATIONAL ACADEMY OF SCIENCES OF THE UNITED STATES OF AMERICA 50, 577 (2007).
- The crystal structure of the Escherichia coli AmtB-GlnK complex reveals how GlnK regulates the ammonia channel
PROCEEDINGS OF THE NATIONAL ACADEMY OF SCIENCES OF THE UNITED STATES OF AMERICA 104, 1213 (2007).DOI: 10.1073/pnas.0610348104
2006
- Structural and functional insights into the AmtB/Mep/Rh protein family
TRANSFUSION CLINIQUE ET BIOLOGIQUE 13, 65 (2006).DOI: 10.1016/j.tracli.2006.02.014