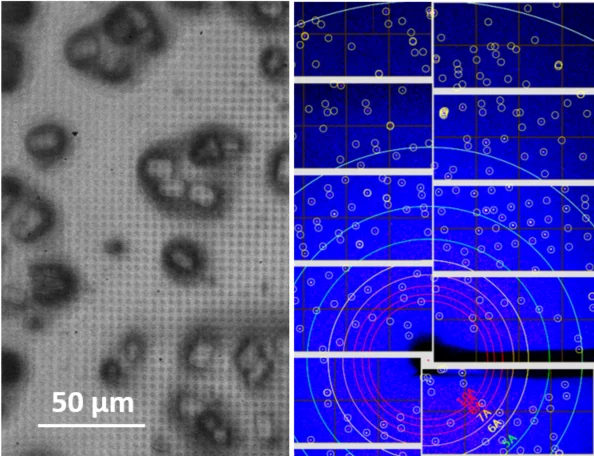
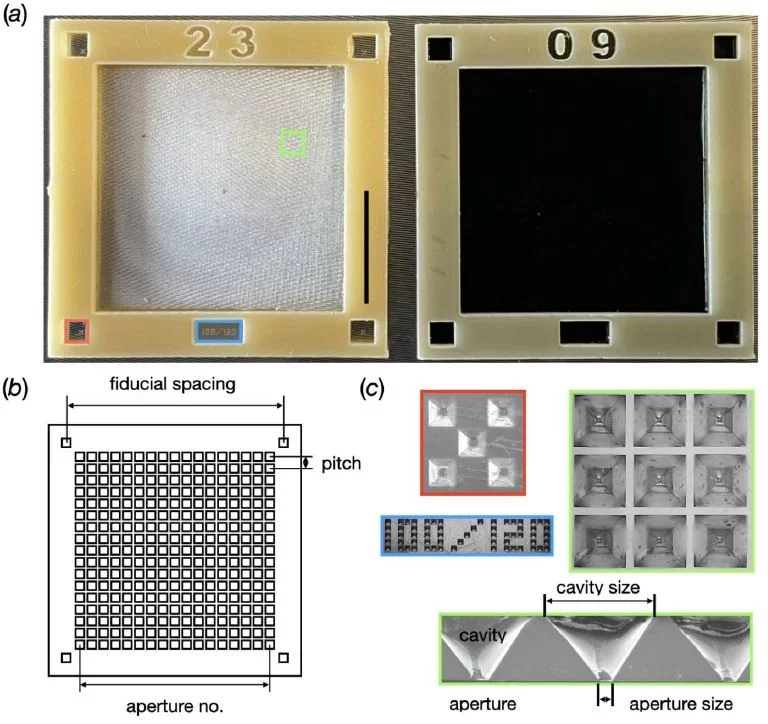
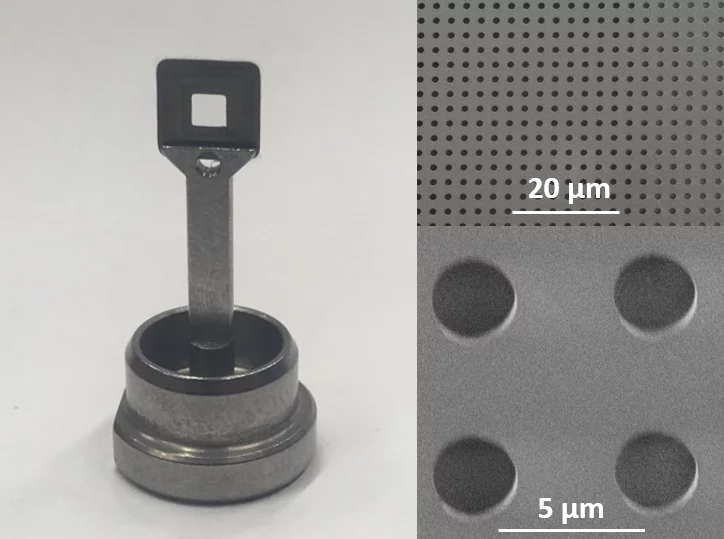
In close collaboration with the macromolecular crystallography group at PSI and the Institute of Polymer Nanotechnology at PSI/FHNW, we are developing solid supports for serial protein crystallography at the SLS and at SwissFEL [1-3].
Serial femtosecond crystallography (SFX) is a powerful new method for protein structure determination at X-ray free electron lasers (XFELs) and synchrotron X-ray sources [4, 5]. It is based on collecting diffraction patterns from large numbers of small protein crystals and allows solving protein structures directly from protein microcrystals, which are too small for standard X-ray techniques. Furthermore, the femtosecond short duration of XFEL pulses allows for dynamic studies of fast structural changes in proteins using pump-probe techniques [6].
To deliver the protein microcrystals at high frequency to the probing X-ray beam, novel protein crystal handling methods such as liquid jet and viscous media injection technologies, as well as the so-called fixed target technology have been developed [7, 8]. In the latter approach, the crystalline sample is deposited on a thin film support, which is mounted on a scanning stage and scanned through the beam, thus sequentially probing the individual microcrystals with the tightly focused x-rays.
The supports are usually fine grid structures or microporous membranes, traditionally produced using silicon/silicon nitride technology, onto which the protein crystal suspension is deposited and separated from the mother liquid through their sieve-function.
We are focusing on the development and fabrication of polymer-based devices, thus taking advantage of the low X-ray absorption and scattering background of polymer materials, absence of X-ray diffraction if using amorphous polymers, the high design flexibility and the potential mass-fabrication at low cost.
MISP-Chips
(Microstructured Polymer Chips) [1]
- developed for RT-serial protein crystallography at SwissFEL/CristallinaMX
- apperture-alignment of crystals
- Small – 6,000 apertures
- Large – 29,000 apertures
- Aperture pitch = 120 µm
- Typically sealed in two layers of Mylar film
standard 6 µm, but thinner films are possible. - Suitable for SFX and SFX pump-probe
SwissMX supports [2,3]
- developed for serial protein crystallography at XFELs and synchrotrons
- minimized background
- efficient blotting
- solutions for cryo and RT measurements
- optimized for use at the SwissMX instrument at SwissFEL
- adaptable to other formats
- fabrication based on planar technology and additive manufacturing
For more information and test samples
please contact Celestino Padeste.
References:
[1] M. Carrillo et a., Micro-structured polymer fixed targets for serial crystallography at synchrotrons and XFELs. IUCrJ (2023) 10, 678-693 doi: 10.1107/S2052252523007595.
[2] A. Karpik, I. Martiel, P.M. Kristiansen, C. Padeste, Fabrication of ultrathin suspended polymer membranes as supports for serial protein crystallography, Micro and Nano Engineering. 2020; 7: 100053 (6 pp.).
[3] I. Martiel et al., Versatile microporous polymer-based supports for serial macromolecular crystallography, Acta Cryst. (2021). D77, 1153–1167, doi: 10.1107/S2059798321007324.
[4] S. Boutet et. al., High-resolution protein structure determination by serial femtosecond crystallography. Science 337 (2012) Vol. 6092, pp. 362-364.
[5] T. Weinert et al., Serial millisecond crystallography for routine room-temperature structure determination at synchrotrons, Nat. Commun. 8, 542 (2017).
[6] V. Paneels et al., Time-resolved structural studies with serial crystallography: A new light on retinal proteins. Structural Dynamics, 2, 041718 (2015)
[7 I. Martiel, H.M. Müller-Werkmeister & A.E. Cohen, Strategies for sample delivery for femtosecond crystallography. Acta Crystallogr. Sect. D. 75, 160–177 (2019).
[8] M.S. Hunter et al., Fixed-target protein serial microcystallography with an x-ray free electron laser, Sci. Rep. 4, 6026 (2014).