All living organisms, from bacteria to humans, rely on proteins to perform their vital functions. How these proteins accomplish their tasks depends on their structure. Researchers from the Paul Scherrer Institute have now devised a novel method to determine the crystal structure of proteins using X-ray light, which could also hasten the development of new drugs in future. The study will be published in the journal Nature Methods on 15 December.
The wavelength of light plays a key role in the depiction of an object, because only details with dimensions that are roughly as big as the wavelength of the light can be resolved. To resolve the structure of a molecule such as a protein on an atomic scale, one needs light with a very short wavelength: in other words, X-ray light.
From the DNA double helix to the present
Ever since the famous double helix structure of a DNA molecule was successfully deciphered using X-ray light in the early 1950s, the capabilities of this method have been greatly exploited. The vast majority of protein structures known today have been determined by X-ray crystallography. The structure of the protein creates a unique diffraction pattern when it interacts with X-ray light. The crystal structure of the protein can be derived from the diffraction pattern, which is recorded on a detector located behind the sample. Thanks to the high quality X-ray light generated at synchrotron light sources, protein crystallography is now highly efficient. One of the top synchrotron light sources in the world is PSI’s Swiss Light Source, where the new technique was developed.
From niche application to new standard
PSI researchers improved a method known as “native SAD” (single-wavelength anomalous diffraction), which was first used in the early 1980s. A typical structure determination of a new protein depends on the incorporation of heavy, exogenous atoms, which is a tedious step in most cases. Here, the native SAD method only uses atoms present in the protein, such as sulphur, to determine the structure. Until now, however, this method was limited to the structures of small proteins.
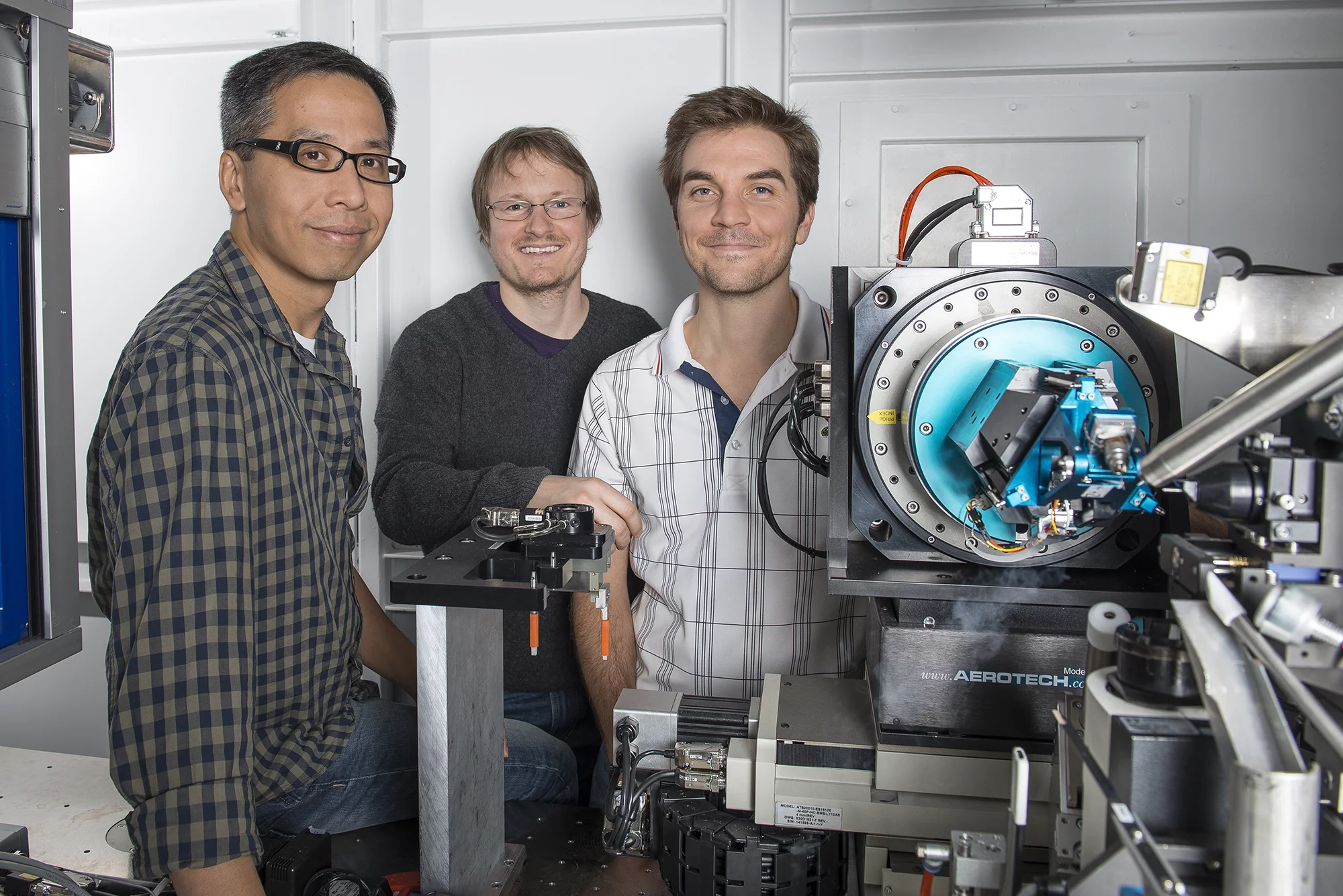
The high sensitivity of the PILATUS detectors produced by Dectris, a spin-off company of PSI, was important for the technique’s success. “These detectors can record signals with low noise with a low intensity of X-ray light, which means one can determine the protein structure with a low X-ray dose,” explains PSI scientist Vincent Olieric. Using conventional techniques, too high a dose of radiation is often necessary to obtain a diffraction image of the protein in the first place, which in some cases can damage the protein to the point that the structure cannot be determined.
In order to be able to collect data of sufficiently good quality and accuracy with low radiation dose, the researchers resorted to another trick. Not only did they rotate the sample on a single axis as usual, they also rotated the sample along two other axes, using the PRIGo multi-axis goniometer developed at PSI. The major advancement of the PRIGo design was to reduce the size of the goniometer, while still maintaining the necessary high precision, in order to avoid collision with other parts of the setup- a major problem at most beamlines.
The technique passes the test
“Using our technique, we managed to determine over twenty protein structures in the space of thirty months. In the last two decades, only about a hundred structures have been solved with the native SAD method. This shows how our method can speed up the entire process as it works without modifications to the protein,” says Tobias Weinert, the first author of the study. The protein structures that the PSI researchers have elucidated with their new technique include that of a “tubulin” molecule (provided by the research group of Michel Steinmetz at PSI), which strengthens the skeletons of many cells. “This is the largest and most complex protein structure solved with the native SAD technique so far. Before our study, virtually everyone thought it was impossible to determine this protein structure in its original state with native SAD.”
As most proteins are smaller and less complex than tubulin, this means that many more structures can now be solved with this improved method, elevating it to a new standard and thus rendering the structural determination of many proteins quicker, simpler, and more cost-effective in the future.
Text: Paul Scherrer Institute/Leonid Leiva
About PSI
The Paul Scherrer Institute PSI develops, builds and operates large, complex research facilities and makes them available to the national and international research community. The institute's own key research priorities are in the fields of matter and materials, energy and environment and human health. PSI is committed to the training of future generations. Therefore about one quarter of our staff are post-docs, post-graduates or apprentices. Altogether PSI employs 1900 people, thus being the largest research institute in Switzerland. The annual budget amounts to approximately CHF 350 million.
Additional information
Macromolecular Christallography GroupContact
Dr. Meitian Wang, Leader of the Group Macromolecular Christallography, Paul Scherrer Institute,Telephone: +41 56 310 41 75, E-mail: meitian.wang@psi.ch
Original Publication
Fast native-SAD phasing for routine macromolecular structure determinationTobias Weinert et al.,
Nature Methods 15 December 2014, Advance Online Publication
DOI: 10.1038/nmeth.3211