Hot vents in the deep sea are home to microbes that feed on ethane. They were discovered recently from scientists of the Max Planck Institute for Marine Microbiology. Now the researchers from Bremen succeeded in finding an important component in the microbial conversion of the gas. They were able to decode the structure of the enzyme responsible for the ethane fixation. The structure highlights some common traits with the methane-fixing counterpart, but also revealed the key features for ethane specialization. The results have now been published in the renowned journal Science.
This insight is the result of the close collaboration of several research groups at the Max Planck Institute for Marine Microbiology. A team around Cedric Hahn and Gunter Wegener recently discovered ethane-degrading microbes at hydrothermal vents of the Guaymas Basin at a water depth of 2,000 meters in the Gulf of California. They named it Ethanoperedens thermophilum, which means “heat-loving ethane-eater”. Cedric Hahn a PhD student from the research group Molecular Ecology cultured the ethane-degrading microbes in the laboratory. Hahn, Wegener and colleagues of the research group Microbial Metabolism, Tristan Wagner and Olivier Lemaire took a closer look at these microorganisms. This collaborative work unraveled the secrets behind the ethane fixation. “We were astonished by what we found. Besides a global similarity, some features of the enzyme differ fundamentally from its counterpart – the enzyme responsible for the degradation of methane,” says Gunter Wegener, scientist in the Research Group for Deep-Sea Ecology and Technology.
Ethane-eaters depend on the same enzyme as Methane-eaters
In deep-sea sediments, geothermal heat leads to the degradation of organic matter to oil and natural gases such as ethane. The ethane is consumed by different microorganisms that form a so-called consortium: archaea, which break down the natural gas, and bacteria, which couple the electrons released in the process to the reduction of sulfate, an abundant compound in the ocean. The discovery of the ethane-eating microbes has brought a breath of fresh air to the research. Compared to microbes eating methane that take a lot of time to grow, the ethane specialists grow much faster and double every week. Thus, the time of biomass production is reduced, which allow attempts of purification and characterization of key enzymes catalyzing the oxidation of natural gas.
To test similarities between the enzymes catalyzing the activation of ethane and methane, Cedric Hahn added a well-known molecular inhibitor of methane oxidation to his culture. This treatment abolished also ethane oxidation. “This suggested that the ethane-oxidizing archaea activate ethane in similar enzymatic reactions as those acting in methane degradation/generation” says Cedric Hahn. Such enzymes are a key expertise of Tristan Wagner who studied them for several years.
A structure visualized at an astonishing level of precision
Cedric Hahn and Olivier Lemaire, the two first authors of the paper now published in Science, then tried to purify the enzyme responsible for ethane fixation. “The project was highly challenging,” says Olivier Lemaire. “Usually we purify the enzymes from much larger amounts of biomass from a culture containing one microorganism. However, we finally yielded sufficient amounts of pure enzymes for structural analyses.”
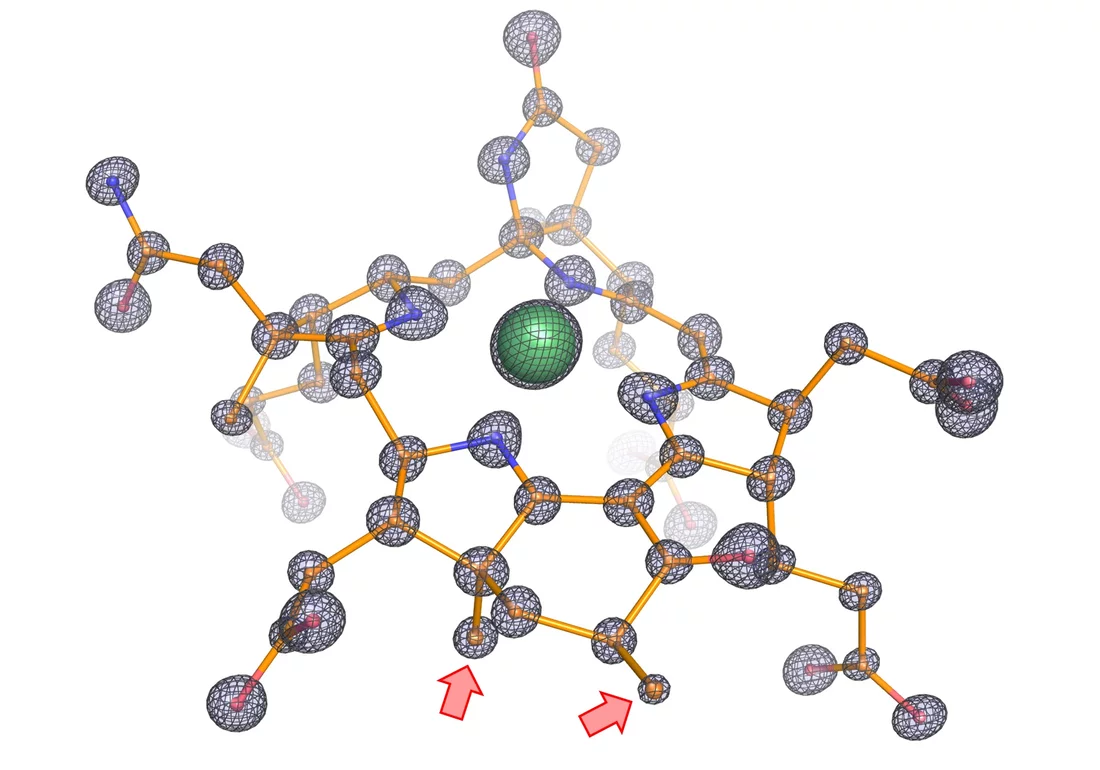
The next critical step was to obtain crystals of the enzyme to determine its tri-dimensional structure. “X-ray crystallography gave previous excellent results on this group of enzymes” says Tristan Wagner, head of the Microbial Metabolism research group and expert on this technique. “We analyzed these crystals by X-ray diffraction and solved the enzyme structure at an unprecedented atomic resolution. We can thus determine the position of individual atoms and obtain therefore an extremely precise picture of the structure”. Beside others they used the beamline X06DA (PXIII) at the Swiss Light Source SLS.
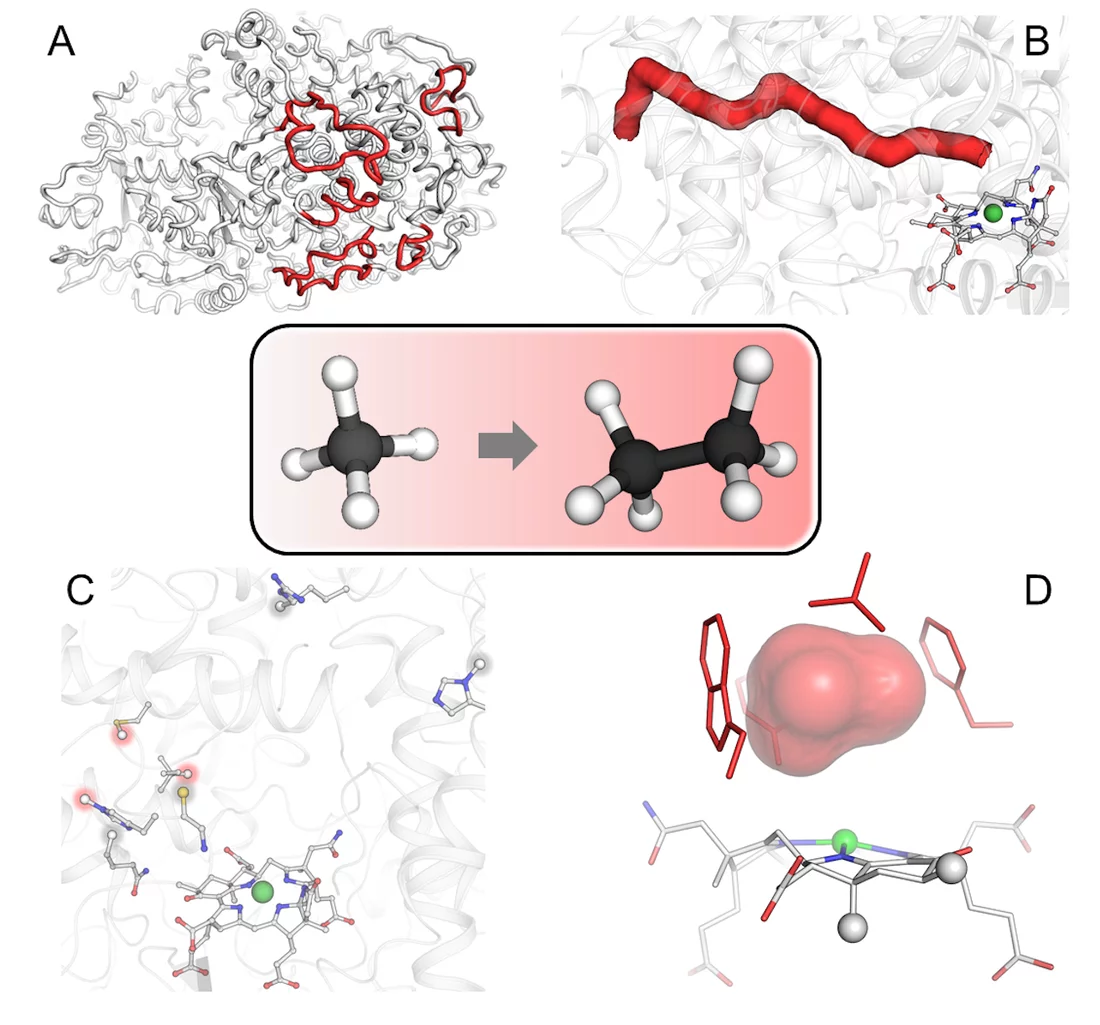
The structure exhibits several unprecedented features. “We noticed that the catalytic chamber in which the chemical reaction takes place is twice as large as in enzymes capturing methane, which makes sense considering that ethane is bigger than methane,” says Olivier Lemaire. The cofactor, catalyst of the reaction, contains two additional methyl groups. These methyl groups were confirmed by Jörg Kahnt from the Max Planck Institute for Terrestrial Microbiology, a worldwide expert on this cofactor. “We found a protein that could be responsible for these methylations, and it is only found in ethane-consumers,” says Cedric Hahn. Since the chamber is more voluminous, a normal cofactor would simply not fit properly and would impair the reaction. The methylations on the cofactor anchor it in the correct position.
Furthermore, the enzyme contains a tunnel connecting the exterior to the catalytic chamber. This tunnel does not exist in any characterized similar enzymes. The researchers experimentally proved the existence of this tunnel through a collaboration with Sylvain Engilberge at the Paul Scherrer Institute in Switzerland, where the protein crystals were gassed with xenon. Xenon was detected in the catalytic chamber and the predicted gas tunnel, proving its existence. The tunnel is maintained and stabilized by modified amino acids and additional extensions.
Now the spotlight turns to propane and butane
The enzyme structure illustrates how these microbes from geothermally active seeps became specialized in ethane capture. This work allows a deeper understanding of the first step in ethane degradation, the only source of energy of these archaea. “Our finding that the enzyme responsible for the process has specific traits to recognize ethane rather than other alkanes is a big step forward, however the understanding of the whole degradation process is still a long way off”, concludes Tristan Wagner.
So, how to proceed the research? “Our previous works show that the activation of longer alkanes requires similar enzymes” says Gunter Wegener. “As a next step, we want to investigate what could be the specific features of the enzymes that catalyze the activation of propane and butane”.
Text based on a press release of Max-Planck-Institute for marine Microbiology
Original Publication
Crystal structure of a key enzyme for anaerobic ethane activation
Cedric J. Hahn*, Olivier N. Lemaire*, Jörg Kahnt, Sylvain Engilberge, Gunter Wegener, Tristan Wagner
Science, July 2021,
DOI: 10.1126/science.abg1765
*Both authors contributed equall
Participating institutions
- Max Planck Institute for Marine Microbiology, Bremen, Germany
- Max Planck Institute for Terrestrial Microbiology, Marburg, Germany
- Paul Scherrer Institute (PSI), Villigen, Switzerland
- MARUM – Center for Marine Environmental Sciences, Bremen, Germany
Please direct your queries to:
Cedric Hahn
Department of Molecular Ecology
Max Planck Institute for Marine Microbiology
Phone: +49 421 2028-9670, e-mail: chahn@mpi-bremen.de
Olivier Lemaire
Research Group Microbial Metabolism
Max Planck Institute for Marine Microbiology
Phone: +49 421 2028-7460, e-mail: olemaire@mpi-bremen.de
Dr. Tristan Wagner
Head of the Research Group Microbial Metabolism
Max Planck Institute for Marine Microbiology
Phone: +49 421 2028-7440, e-mail: twagner@mpi-bremen.de
Dr. Gunter Wegener
Research Group Deep-Sea Ecology and Technology
Max Planck Institute for Marine Microbiology
Phone: +49 421 2028-8670, e-mail: gwegener@mpi-bremen.de
Katrin Matthes
Press Officer
Max Planck Institute for Marine Microbiology
Phone: +49 421 2028-9470, e-mail: presse@mpi-bremen.de