Molecular basis for the development of new drugs against autoimmune diseases
Like a shredder, the immunoproteasome cuts down proteins into peptides that are subsequently presented on the cellular surface. The immune system can distinguish between self and non-self peptides and selectively kills cells that, due to a viral infection, present non-self peptides at their surface. In autoimmune diseases, this mechanism is deregulated and the immune system also eliminates uninfected cells by mistake. However, inhibition of the immunoproteasome may alleviate disease symptoms and progression. Biochemists at the Technische Universitaet Muenchen (TUM), with the help of measurements taken at the Paul Scherer Institute, have now succeeded in determining the first crystal structure of an immunoproteasome. The results are reported in the renowned journal Cell and will enable the development of new drugs that selectively target the immunoproteasome.
The proteasome, a large, cylindrical protein complex, plays a key cellular role. Like a recycling facility, it breaks down superfluous proteins into smaller fragments that can subsequently be reused. Hence, proteasomes regulate essential cell functions, such as cell survival, growth and division, as well as the removal of defective proteins.
Similar and yet different: Damaged balance between molecules leads to illness
Apart from the normal, so-called constitutive
proteasome, mammals also bear a specialized version, the immunoproteasome, which differs in a few details from normal, so-called constitutive
proteasome. As an important partner of the immune system, the immunoproteasome breaks the proteins down into pieces, so-called antigens, that can be presented at the cellular surface. Pieces of foreign, e.g. viral, proteins trigger an immune response, ultimately leading to the elimination of infected cells by the immune system.
Structure explained – One building block makes all the difference
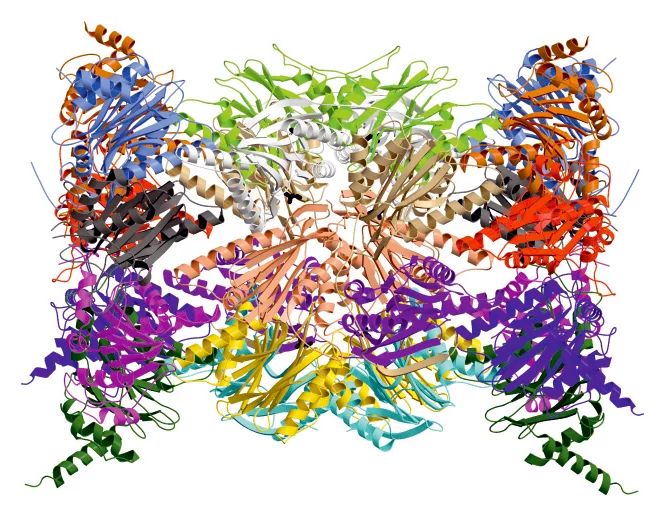
Professor Michael Groll, Chair of Biochemistry in the Department of Chemistry at the TU Muenchen and his co-worker, Eva Maria Huber, in collaboration with Professor Marcus Groettrup, Chair of Immunology at the University of Konstanz (Germany) and Director of the Biotechnology Institute Thurgau (BITg) in Kreuzlingen (Switzerland), have now achieved a milestone: Using X-ray crystallography, they have elucidated the crystal structures of both the constitutive proteasome and the immunoproteasome of mice.
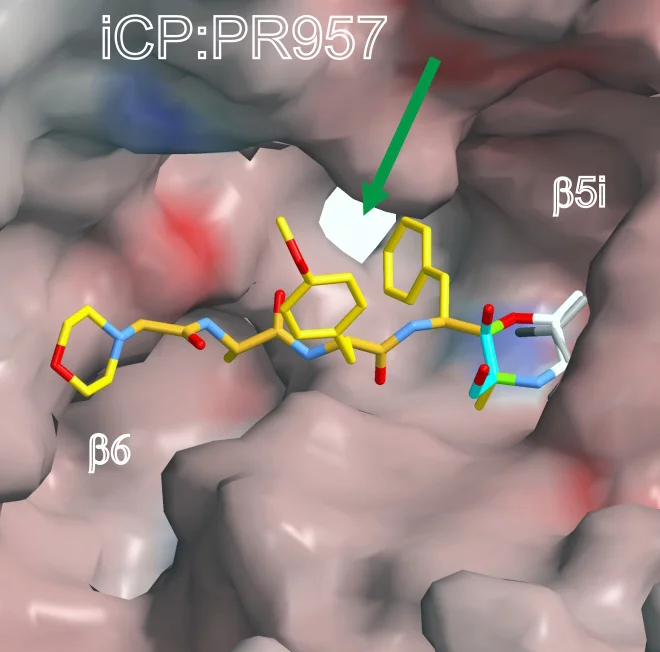
The scientists determined both structures alone and in complex with the compound PR-957 (ONX 0914). PR-957 is a promising proteasome inhibitor that specifically blocks the immunoproteasome but not the constitutive one . However, the molecular reason underlying its selectivity for the immunoproteasome remained unknown. By visualizing the binding of the inhibitor to both proteasome types on an atomic level, we unravelled the molecular basis for the selectivity of PR-957
explains Groll. Based on these results, we now can develop new and even more specific inhibitors.
The crystal structures of the constitutive proteasome and immunoproteasome enabled Groll and Huber to identify the prerequisites for the high affinity of PR-957 only for the immunoproteasome. Remarkably, the orientation of a single amino acid side-chain in the substrate binding pocket of the active site is the key to the increased affinity. Although the substrate binding pockets of both proteasome types are lined with almost identically ordered amino acids, subtle changes in the immunoproteasome cause one particular methionine to adopt a different orientation than in the constitutive proteasome. This distinct conformation is crucial,
emphasizes Eva Maria Huber. It results in a larger pocket in the immunoproteasome, which therefore preferentially accommodates bulky amino acids and also the inhibitor. In contrast, constitutive proteasomes harbour a significantly smaller cavity that hampers binding of PR-957.
Hope for new drugs
The elucidation of the atomic differences between immuno- and constitutive proteasomes represents an important step for the development of new drugs for the treatment of inflammatory disorders, which have often been linked to elevated levels of immunoproteasomes. The insights gained from the X-ray structures will support the development of new compounds that selectively target the immunoproteasome, without affecting the constitutive counterpart, whose proper function is essential for cellular survival.
Synchrotron Light shows structures in detail
The molecular structures for this work were determined at the Swiss Light Source, SLS, at PSI. In the process used, the molecules that were to be studied were first of all arranged into a regular crystal structure and then illuminated with synchrotron light from the SLS. From the way in which the light was subsequently defelcted, researchers were able to determine the exact structure of the molecule. PSI offers three measurement stations at the SLS for this procedure. Researchers from science and industry, from abroad as well as from within Switzerland proceed under great competition to obtain access and consequently measurement time at these measurement stations.
The work was funded by the German Research Foundation (DFG) (SFB595/TP A11), the German Federal Ministry for Education and Research (ProNet-T3/TP To-03), the Swiss National Science Foundation (SNSF) (31003A_138451) and the Excellence Cluster Center for Integrated Protein Science Munich (CIPSM). Measurements were performed at the PXI und PXIII beamlines of the Paul Scherrer Institute (Villigen, Switzerland).
Text based on a press release published by the Technische Universität München
About PSI
The Paul Scherrer Institute develops, builds and operates large, complex research facilities, and makes them available to the national and international research community. The Institute's own key research priorities are in the investigation of matter and material, energy and the environment; and human health. PSI is Switzerland's largest research institution, with 1400 members of staff and an annual budget of approximately 300 million CHF.
Contact
Prof. Dr. Michael Groll, Technische Universität München, Chair of Biochemistry
Lichtenbergstr. 4, 85748 Garching, Germany
Tel: +49 (0)89 289 13361; E-Mail: michael.groll@ch.tum.de
Internet: http://www.biochemie.ch.tum.de/
Prof. Dr. Marcus Groettrup; University of Konstanz; Chair of Immunology
Universitätsstrasse 10; 78464 Konstanz; Germany; Tel: +49 (0)7531 / 88-2254
E-Mail: Marcus.Groettrup@uni-konstanz.de
Dr. Michael Basler, Biotechnologie Institut Thurgau (BITg), Unterseestrasse 47,
8280 Kreuzlingen, Switzerland; Tel: +41 (0)71 678 5020
E-Mail: Michael.Basler@uni-konstanz.de; http://www.bitg.ch
Dr. Takashi Tomizaki, Laboratory for Macromolecules and Bioimaging, Paul Scherrer Institut, Villigen PSI, Switzerland,
Telefon: +41 56 310 51 29, E-Mail: takashi.tomizaki@psi.ch
Original publication
Immuno- and constitutive proteasome crystal structures reveal differences in substrate and inhibitor specificity, Eva M. Huber, Michael Basler, Ricarda Schwab, Wolfgang Heinemeyer, Christopher J. Kirk, Marcus Groettrup, Michael Groll, Cell, 17 February 2012
Additional Information:
Research using synchrotron light
The beamlines used in the experiments:
http://www.psi.ch/sls/pxi/pxi
http://www.psi.ch/sls/pxiii/pxiii