A dream drug would provide a targeted therapeutic effect without side effects. Biased signalling, whereby certain cellular signalling pathways are favoured over others, could make this a reality. Now, publishing in PNAS, PSI researchers have created a platform for biased signalling-based drug discovery. Using their platform, they test specially developed bivalent ligands that induce dimerization of G protein-coupled receptors and observe how they can drive cellular signalling outcomes in different directions.
G protein-coupled receptors (GPCRs), proteins formed from seven helices that span the cell membrane, are the cell’s way of responding to the outside world. The binding of a ligand to the receptor results in structural changes leading to the recruitment of adaptor proteins and the activation of subsequent signalling cascades that mediate cellular responses. GPCRs, in which PSI has world leading research expertise, are one of the largest and most diverse family of membrane receptors in humans. Due to their crucial role in physiology, they have become one of the most important pharmaceutical drug targets with around 25% of prescribed drugs believed to act through this family of proteins.
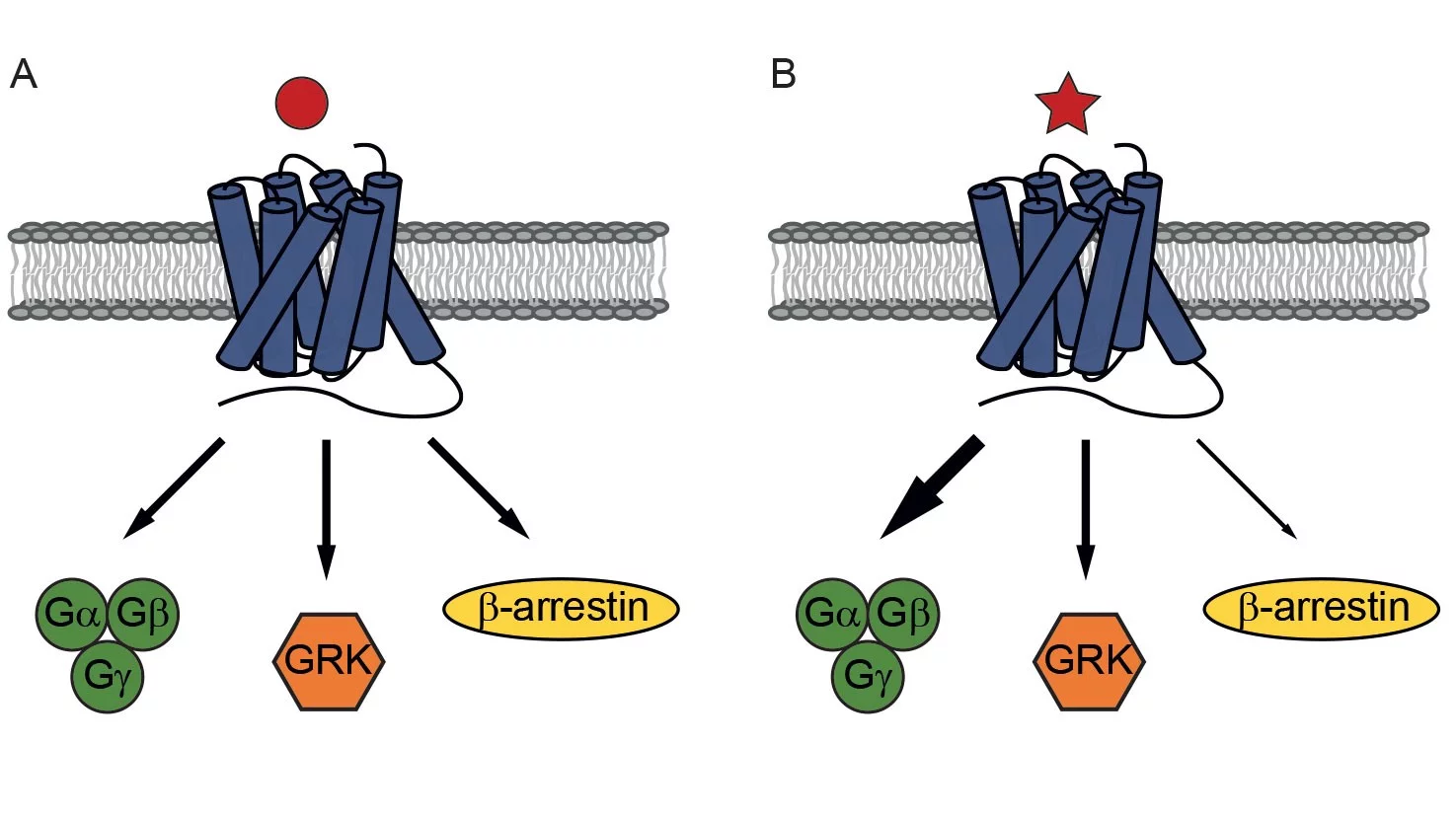
We now know that it is possible to preferentially trigger certain cellular signalling pathways in a phenomenon known as biased signalling. This signalling specificity ultimately originates from the different conformations that ligands can induce when they bind to receptors. This leads to different ratios of recruited adaptor proteins, such as G-proteins, G protein-coupled receptor kinases (GRKs), or β-arrestins, resulting in different signalling pathways and different biological outputs.
“Biased signalling is an exciting prospect for pharmacy, as it gives the ability to favour the desired drug action over unwanted side effects,” explains corresponding author, PSI’s Philipp Berger.
For example, the µ-opioid receptor is the target for many pain medications. Binding of an analgesic, such as morphine, triggers the recruitment of G-proteins, which begins a signalling cascade resulting in pain relief. However, it also triggers the recruitment of β-arrestins, which is linked to undesired effects such as addiction. The ability to bias signalling towards the G-protein recruitment mediated pathway would be a pharmaceutical revolution.
A new platform to measure biased signalling
A challenge for the pharmaceutical development of biased signalling is how the resulting signalling response is measured. Researchers from PSI’s Laboratory for Nanoscale Biology have developed a platform that enables the precise characterisation of ligand-induced bias and, hence, the screening of drug molecules based on signalling response. Using chemiluminescence techniques - namely bioluminescence resonance energy transfer (BRET) and enzyme complementation - to measure conformational changes in the GPCRs, the platform enables the time-resolved recruitment of adaptor proteins in response to ligand binding to be studied. This enables the selectivity of drugs towards the G protein, GRK, and β-arrestin pathways to be explored.
Manipulating the signalling bias with drugs
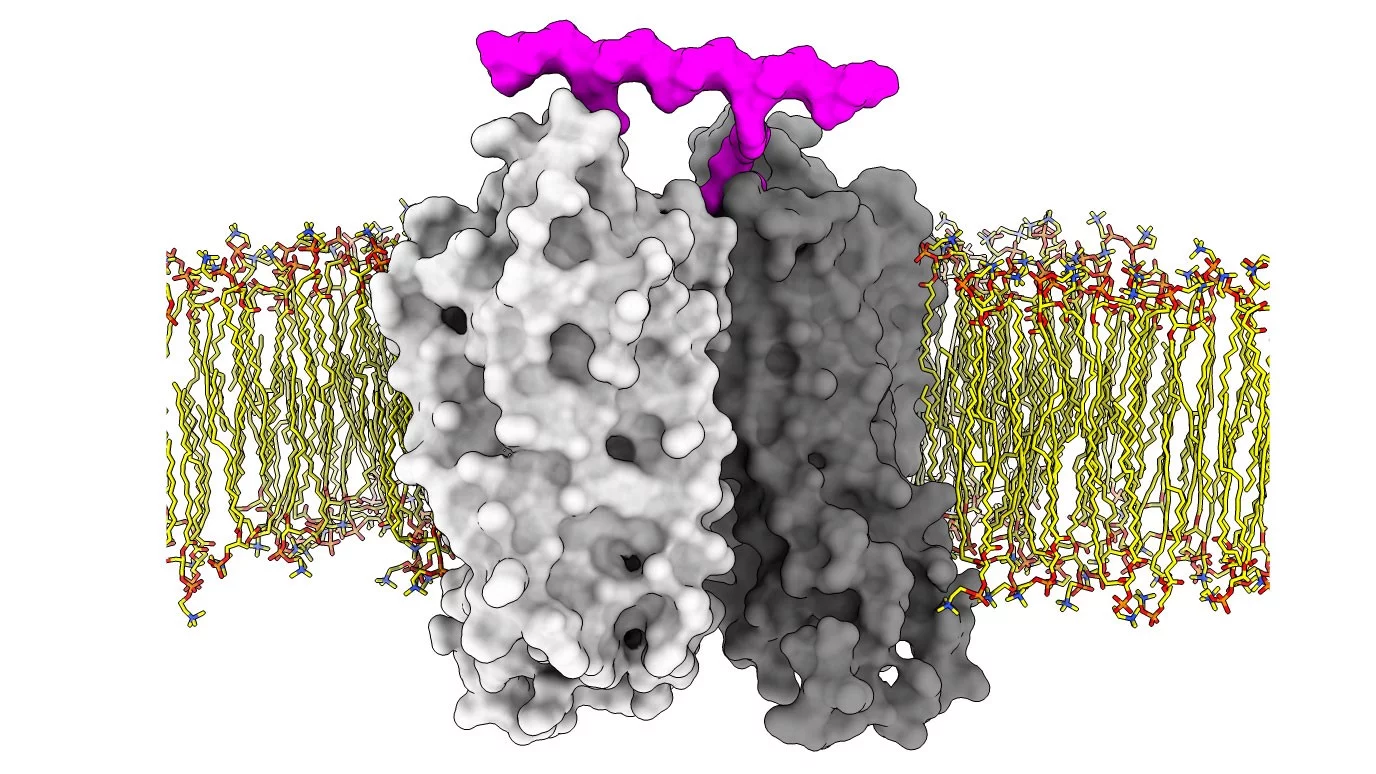
Using their platform, the team tested their ability to manipulate the signalling of GPCRs with drugs. Working together with the Center for Radiopharmaceutical Sciences at PSI and the Laboratory of Organic Chemistry at ETH Zürich, the team designed and created bivalent ligands based on a rigid oligoproline backbone with a fixed distance between two recognition motifs. These can simultaneously dock onto two GPCRs and promote their dimerization. Testing their ligands on the gastrin-releasing peptide receptor, a GPCR found in the gut, brain and certain tumours, the team discovered that inducing dimerization is a means to alter the signalling profile of the receptor. Furthermore, they found that, by varying the distance between the recognition motifs, they could bias signalling towards G-protein recruitment or β-arrestin recruitment, offering a further degree of control.
This work demonstrates the capabilities of the platform to measure drug induced signalling bias. “The platform, suitable for all GPCRs, is highly flexible and provides an exciting opportunity to screen newly synthesised drugs based on their signalling preference,” says Philipp Berger.
In the future, the ability to screen drugs based on their signalling profile could lead to the development of drugs with more selective therapeutic effect and fewer side effects.
Text: Miriam Arrell / Paul Scherrer Institute
Contact
Dr. Philipp Berger
Paul Scherrer Institute
Forschungsstrasse 111
5232 Villigen PSI
Switzerland
Telefon: +41 56 310 47 28
E-Mail: philipp.berger@psi.ch
Original Publication
Exploring the signaling space of a GPCR using bivalent ligands with a rigid oligoproline backbone
N. Romantini, S. Alam, S. Dobitz, M. Spillmann, M. De Foresta, R. Schibli, G.F. X. Schertler, H. Wennemers, X. Deupi, M. Behe, and P. Berger
PNAS 118 (48)
DOI: 10.1073/pnas.2108776118
Further Information
Mechano-Genomics Group | MGG | Paul Scherrer Institut (PSI)
Schertler Group Research | BIO | Paul Scherrer Institut (PSI)
Wennemers Group | ETH Zürich Laboratory of Organic Chemistry
Funding
This work was supported by a Sinergia grant from the Swiss National Science Foundation (CRSII2_160805) and a National Centre of Competence in Research (NCCR) Molecular Systems Engineering grant.
Copyright
PSI provides image and/or video material free of charge for media coverage of the content of the above text. Use of this material for other purposes is not permitted. This also includes the transfer of the image and video material into databases as well as sale by third parties.